
Viral presence and the bacterial microbiome in chronic rhinosinusitis
Introduction
The bacterial microbiome of the aerodigestive tract and specifically the sinonasal passages is an area receiving increasing attention in current literature. This is of significant interest to research into chronic rhinosinusitis (CRS). This inflammation of the nose and paranasal sinuses of greater than twelve weeks duration is characterised by symptoms of anterior rhinorrhoea, post-nasal drip, headaches, facial pain or pressure, nasal obstruction and hyposmia. Aetiopathogenesis theories are many but a significant focus remains on microbial imbalance, and methods of manipulating this for therapeutic ends.
A bacterial dysbiosis has been shown to exist within CRS with patients showing a lack of biodiversity compared to healthy controls (1). Bacterial species historically seen as more pathogenic are found to be relatively more prevalent in CRS than bacterial species that have been considered commensal (2). Studies investigating the CRS microbiome have varied in scale, methodology and analysis and so data have been difficult to compare. Efforts have been made to standardise these, concluding that the healthy sinonasal microbiome consists mainly of Staphylococcus, Propionibacterium, Corynebacterium and Streptococcus species while the microbiome in CRS shows a reduction in the relative abundance of Actinobacteria and Propionibacterium species, with significantly more prevalent Corynebacterium species (3).
Upper respiratory tract viruses have recently been shown to be more common in CRS than in healthy individuals. The presence of these viruses is also associated with more severe subjective and objective disease (4). These findings potentially implicate viruses as an inciting and/or exacerbating factor in the immune dysregulation of CRS. Viral-bacterial co-infection is known to have deleterious effects on epithelial barrier function, bacterial binding and innate and adaptive immunity (5). It has been suggested that viruses may induce changes in the bacterial microbiome potentially causing more severe disease in the lower respiratory tract (6,7). However, little is known about the specific changes in bacterial aerodigestive populations seen in the presence of viral infection in CRS.
The aim of this study was thus to investigate virus-associated changes in the bacterial CRS sinonasal microbiome, hypothesising that viral infection would indeed alter the composition of resident bacteria. We present the following article in accordance with the STROBE reporting checklist (available at https://www.theajo.com/article/view/10.21037/ajo-21-53/rc).
Methods
Study participants
Study participants were recruited from the tertiary rhinologic practices of two of the senior authors (PJW and AJP) in Adelaide, Australia over the course of 2017 and 2018. This study was carried out in accordance with the recommendations of the Central Adelaide Local Health Network Ethics Committee, with their approval of the protocol (HREC/15/TQEH/132). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and study participants gave verbal and written informed consent. Patients were included in this study if they were older than eighteen years of age and were undergoing endoscopic nasal surgery. Control patients did not have any clinical or radiologic evidence of CRS, and were undergoing trans-sphenoidal resections of pituitary masses or surgery to the septum or inferior turbinates. CRS patients fulfilled the diagnostic criteria for CRS as outlined in the American guidelines, and were undergoing functional endoscopic sinus surgery (FESS) (8). Patients who had used antibiotics or steroids in the two months prior to the study day were excluded. No patients in this study suffered from asthma or aspirin-exacerbated respiratory disease and none were smokers. On the study day patients were asked to report the timing of their last URTI with the following time points: current, within the last 1–2 weeks, within the last 2–4 weeks, within the last 1–2 months, or more than 2 months prior.
Viral sampling, processing and analysis
Viral sampling, processing and analysis was undertaken using a previously published departmental protocol (9). Briefly, the left and right middle meatus (MM) and inferior meatus (IM) mucosa was sampled using EndoScan cytology brushes (McFarlane Medical, Melbourne, Australia). This was conducted with endoscopic visualisation and aseptic technique. Samples were transported on ice and stored at −80 °C. At time of processing samples were thawed for RNA and DNA extraction using an AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The products of these were stored and subsequently real-time PCR batch tested. An initial assay for endogenous retrovirus 3 (ERV3) was undertaken in the DNA extract fractions to ensure adequate sample collection quality. Assays were then undertaken for adenovirus (AdV), bocavirus (BoV), coronavirus (CoV), enterovirus (EnV), influenza, metapneumovirus (MPV), parainfluenza (PIV), respiratory syncytial virus (RSV) and rhinovirus (RV). PCR target genes, primer and probe sequences, the nature of positive and negative controls and cycling conditions have been published previously (9). A cycle threshold (Ct) of forty or less indicated viral detection, as has been validated previously (9).
Bacterial sampling and processing
Bacterial sampling was also undertaken intra-operatively with an aseptic technique and endoscopic visualisation. Guarded, flocked swabs (Copan Italia S.p.A, Brescia, Italy) were inserted into the MMs of all patients on both sides and rotated seven times before removal. Swabs were stored at −80 °C until batch thawing for DNA extraction and analysis as follows. Swab heads were cut into small pieces and 180 µL of enzymatic lysis buffer (Qiagen, Hilden, Germany) was added and left overnight at room temperature. 5 mm steel beads agitated for 20 seconds at 15 Hz in a Qiagen Tissue Lyser were used to homogenise the pieces, followed by 5 minutes of further homogenisation with 0.1 mm glass beads at 30 Hz. DNA extraction was then undertaken in accordance with the Qiagen DNeasy Blood and Tissue Kit protocol (Qiagen, Hilden, Germany), and stored at −80 °C until sequencing.
Bacterial 16S-sequencing
Polymerase chain reaction (PCR) amplification and sequencing was performed by the Australian Genomics Research Facility (AGRF). Gene libraries were generated by amplifying the V3 to V4 (341F–806R) hypervariable region of the 16S rRNA. AmpliTaq Gold 360 Master Mix (Life Technologies, Mulgrave, Australia) with primers CCTAYGGGRBGCASCAG in the forward sequence and GGACTACNNGGGTATCTAAT in the reverse sequence were used to generate PCR amplicons. These underwent fluorometric measurement (Invitrogen Picogreen; Thermo Fisher Scientific, Waltham, MA, USA) and normalised. Quantitative PCR (KAPA Biosystems, Capetown, South Africa) was used to quantify the equimolar pool. This was arranged for sequencing on the Illumina MiSeq (Illumina Inc., San Diego, CA, USA) with 300 base paired end chemistry. All samples in this study were sequenced in one run.
Statistical analysis and bioinformatic pipeline
The Quantitative Insights Into Microbial Ecology platform (QIIME 2, version 2018.11) (10) was employed for the bioinformatic pipeline in this study. PEAR was used to combine forward and reverse reads (11) through the QIIME 2 plugin q2-pear (https://github.com/bassio/q2-pear). The QIIME 2 plugin q2-quality-filter (12) was used to quality-filter the combined sequences. Minimum quality parameter was 20 (13). Deblur (q2-deblur plugin with setting “trim-size” =435 with default parameters otherwise) was used for denoising and to form Amplicon Sequence Variant (ASV) (13). Greengenes 16S database (version 13.8 August 2013, the 99% clustered similarity sequences) was used as reference (14). and the QIIME 2 BLAST-based q2-feature-classifier was used as taxonomy classifier (15).
Prior to analysis, n=400 reads were chosen as the rarefaction depth cut-off. Rarefaction plots (for total number of ASVs and for Shannon’s alpha diversity index) were performed (Figure S1) and showed that most samples had rarefaction curves that reached a plateau at the 400 read depth, indicating sufficient subsampling. Relative abundance comparisons were done at the genus level. The taxonomic assignment of the one DNA-negative control sample containing extraction reagents only was explored. The bacterial genus Flavobacterium was present in high relative abundance in this sample and in relatively low abundance in many samples, so this genus was excluded before downstream statistical analyses. Mean relative abundance and genera prevalence were calculated. Alpha diversity was measured using Shannon’s diversity and Faith’s phylogenetic diversity index (16), calculated using Sci-kit bio (version 0.5.3).
Rank variability is a per-sample index, and a surrogate for microbiome stability. It is defined by Martí et al. as “the absolute difference between each taxon rank and the overall rank” (17). Rank variability was calculated using a Python implementation of the equations previously described (17).
Results
Patient characteristics
A total of 82 patients were recruited: 10 controls, 49 CRSsNP and 23 CRSwNP. Of 82 patients, 41 were virus-positive; 5 control, 24 CRSsNP and 12 CRSwNP. Virus-positive and virus-negative patients were age and season-matched within the three groups (control, CRSsNP and CRSwNP). Demographics and patient characteristics are summarised in Table 1.
Table 1
| Demographics | Control | CRSsNP | CRSwNP |
|---|---|---|---|
| Number with each diagnosis | 10 | 49 | 23 |
| Number of virus-positive patients | 5 | 24 | 12 |
| Mean age (years) | 35.5 | 50.0 | 45.0 |
| Male:female | 4:6 | 27:22 | 22:1 |
| Season sample obtained (spring:summer:autumn:winter) | 2:2:2:4 | 20:8:12:9 | 4:4:8:7 |
CRSsNP, CRS without nasal polyps; CRSwNP, CRS with nasal polyps; CRS, chronic rhinosinusitis.
Viral detection
ERV3 was detected in all samples. Mean ERV3 Ct was 22.9, indicating that adequate cellular material was obtained in all cases. Forty-one patients were positive for one or more of the disease-causing viruses assayed, while 41 were virus-negative. RV was the most prevalent of the species assayed; MPV was not detected in any of the samples. Fourteen patients were positive for more than one viral species. Details of viral species detected are summarised in Table 2.
Table 2
| Viral species | Number of positive control patients | Number of positive CRSsNP patients | Number of positive CRSwNP patients |
|---|---|---|---|
| Adenovirus | 1 | 0 | 0 |
| Bocavirus | 1 | 2 | 1 |
| Coronavirus | 4 | 6 | 5 |
| Enterovirus | 1 | 1 | 1 |
| Influenza | 0 | 6 | 1 |
| Parainfluenza | 0 | 1 | 2 |
| Rhinovirus | 1 | 13 | 1 |
| RSV | 0 | 0 | 1 |
CRSsNP, CRS without nasal polyps; CRSwNP, CRS with nasal polyps; CRS, chronic rhinosinusitis; RSV, respiratory syncytial virus.
Bacterial microbiome outcomes: taxonomy
The most abundant taxa are found in Table 3. The genera Corynebacterium, Staphylococcus, Streptococcus, Haemophilus, and Moraxella were the most abundant. We found no significant difference in differential abundance of the five most abundant genera by diagnosis (i.e., control versus CRSsNP versus CRSwNP; Kruskal-Wallis P>0.05, Figure 1).
Table 3
| Bacterial genus | Control | CRSsNP | CRSwNP | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Virus-pos | Virus-neg | Virus-pos | Virus-neg | Virus-pos | Virus-neg | ||||||||||||
| MRA | P | MRA | P | MRA | P | MRA | P | MRA | P | MRA | P | ||||||
| Corynebacterium | 26.00 | 80.0 | 44.85 | 100.0 | 33.39 | 58.33 | 31.71 | 60.87 | 23.75 | 66.67 | 48.05 | 58.33 | |||||
| Staphylococcus | 47.06 | 80.0 | 33.05 | 100.0 | 22.45 | 58.33 | 24.49 | 69.57 | 26.85 | 66.67 | 23.55 | 50.00 | |||||
| Streptococcus | 9.12 | 20.0 | 0.00 | 0.0 | 4.86 | 12.50 | 9.94 | 26.09 | 0.00 | 0.00 | 3.62 | 16.67 | |||||
| Haemophilus | 0.00 | 0.0 | 0.00 | 0.0 | 10.55 | 16.67 | 3.50 | 8.70 | 5.42 | 8.33 | 0.00 | 0.00 | |||||
| Moraxella | 0.94 | 20.0 | 4.35 | 40.0 | 7.18 | 16.67 | 0.00 | 0.00 | 0.00 | 0.00 | 3.68 | 8.33 | |||||
| Proteus | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.00 | 0.00 | 0.00 | 8.38 | 8.33 | 10.00 | 8.33 | |||||
| Porphyromonas | 0.00 | 0.0 | 0.00 | 0.0 | 6.11 | 8.33 | 0.00 | 0.00 | 3.60 | 8.33 | 0.00 | 0.00 | |||||
| Genus unidentified (Enterobacteriaceae) | 1.25 | 20.0 | 0.00 | 0.0 | 3.11 | 12.50 | 4.84 | 4.35 | 0.00 | 0.00 | 0.00 | 0.00 | |||||
| Alloiococcus | 0.00 | 0.0 | 0.90 | 40.0 | 0.00 | 0.00 | 2.96 | 4.35 | 4.20 | 8.33 | 0.95 | 8.33 | |||||
| Genus unidentified (Cytophagaceae) | 0.00 | 0.0 | 5.45 | 40.0 | 1.68 | 25.00 | 1.87 | 17.39 | 0.00 | 0.00 | 0.00 | 0.00 | |||||
CRSsNP, CRS without nasal polyps; CRSwNP, CRS with nasal polyps; pos, positive; neg, negative; MRA, mean relative abundance; P, prevalence (both expressed as percentages); CRS, chronic rhinosinusitis.

Viral covariates as predictors of bacterial taxa abundances
We found no significant association between viral presence or number of viruses detected and the abundances of the aforementioned most abundant bacterial genera (Kruskal-Wallis, P>0.05). Moreover there was no statistically significant association detected between the presence of CoV, influenza or RV and bacterial relative abundances. There was also no difference found in relative abundances of the most abundant bacteria when grouped by time since last patient-reported viral infection (Figure 2).
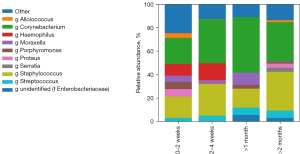
Viral covariates as predictors of bacterial diversity and stability
We calculated Faith’s and Shannon’s indices as markers of both phylogenetic and non-phylogenetic alpha diversity. We also calculated rank variability; a per-sample surrogate for microbiome stability as mentioned earlier. We found no significant association between bacterial genera abundances and viral status covariates such as viral presence, number of viruses, and presence of specific viruses (influenza, CoV or RV) (P>0.05).
Discussion
This study seeks to compare the bacterial microbiome seen in virus-positive and virus-negative individuals both with and without CRS. The most prevalent viruses seen in all groups were CoV, influenza and RV. The most abundant bacterial genera seen were Corynebacterium, Staphylococcus, Streptococcus, Haemophilus, and Moraxella.
No significant differences were seen in bacterial abundance, diversity or stability between virus-negative or virus-positive individuals within the control, CRSsNP or CRSwNP groups with regard to the viral covariates tested (presence or absence of virus, number of viruses, presence of specific viruses or time since last patient-reported viral infection). This is in contrast to previously reported effects of viral presence on the microbiome in non-CRS populations (18-20). Our study focuses on CRS patients, is smaller and geographically different to these studies, but its strengths lie in a robust and previously validated viral collection method (9), an undertaking of more in-depth analysis beyond viral presence or absence alone, and the use of age and season-matched virus-negative control groups. Ding et al. compared swabs from control and influenza-infected individuals (sample size of 40 versus 215). They found Corynebacterium and Streptococcus to be more abundant in controls, with virus-positive swabs dominated by Moraxella and Dolosigranulum (20). This result is interesting not least due to the rarity of identification of Dolosigranulum in adult CRS upper respiratory tract bacterial profiles; the low biomass of airway samples carries risk of contamination if not carefully screened prior to and following analysis. Borges et al. compared swabs from twelve patients with severe acute respiratory infections; six with influenza, and six with undisclosed non-influenza causative viral organisms. Despite their small sample size they found significant differences in abundance of fifteen different bacterial genera, however no control group was presented (19). Rosas-Salazar et al. compared the infantile microbiome in the presence of either RV or RSV, and found significant differences in eleven genera. Again, no control group was presented, and in all three of the aforementioned studies viral collection methods were prone to contamination. No method to ensure cellular collection was employed, and as such even viral presence cannot be confidently asserted in these studies.
The negative effects of a viral/bacterial co-infection have long been established in many body systems, but of significant interest in any such co-infection model is the nature of the original inciting pathogen. This is a cross-sectional study investigating the characteristics of sinonasal microbiota at a solitary time point (that of endoscopic sinonasal surgery). Should differences in these microbiota have been observed between control and CRS patients, we would be unable to determine whether it is the composition of the bacterial microbiome that predisposes to viral infection, or vice versa. To elicit such information a longitudinal study model would be required. To our knowledge this has not yet been undertaken in these cohorts. An additional limitation is a lack of gender balance in the CRSwNP patient sample, and the known differing immune response profile of CRSsNP and CRSwNP patients. CRSsNP is associated with a Th-1 skewed response, while CRSwNP is associated with a Th-2 skewed, eosinophilic response (21,22). Viral infections themselves are also associated with eosinophilia (23). These factors have the potential to impact the detection of virus and/or the microbiome balance observed in this study (24).
Of additional interest but requiring further research would be whether there is correlation between patient reporting of a current viral infection (manifest as more severe symptoms), and the presence of virus in the sinonasal passages at that time. Only three patients in this study reported such, limiting any analyses thereof. An additional limitation is that the symptoms of CRS and of a viral URTI are similar but with a marked difference in duration.
Conclusions
In conclusion this study compares the bacterial microbiome in virus negative and virus positive controls, CRSsNP and CRSwNP, and has uncovered no significant differences in its composition. However, larger, longitudinal investigation is required to investigate this further.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://www.theajo.com/article/view/10.21037/ajo-21-53/rc
Data Sharing Statement: Available at https://www.theajo.com/article/view/10.21037/ajo-21-53/dss
Funding: The study was supported by internal departmental funding only.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://www.theajo.com/article/view/10.21037/ajo-21-53/coif). AJP: Consultant—Medtronic, Fusetec, Tissium, ENT technologies; Speaker Honorarium—Sequiris; Shareholder Chitogel; unpaid editorial board member of Australian Journal of Otolaryngology from January 2021 to December 2022. The other authors have no conflicts of interests to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Central Adelaide Local Health Network Ethics Committee (HREC/15/TQEH/132) and informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Anderson M, Stokken J, Sanford T, et al. A systematic review of the sinonasal microbiome in chronic rhinosinusitis. Am J Rhinol Allergy 2016;30:161-6. [Crossref] [PubMed]
- Psaltis AJ, Wormald PJ. Therapy of Sinonasal Microbiome in CRS: A Critical Approach. Curr Allergy Asthma Rep 2017;17:59. [Crossref] [PubMed]
- Wagner Mackenzie B, Waite DW, Hoggard M, et al. Bacterial community collapse: a meta-analysis of the sinonasal microbiota in chronic rhinosinusitis. Environ Microbiol 2017;19:381-92. [Crossref] [PubMed]
- Goggin RK, Bennett CA, Bialasiewicz S, et al. The presence of virus significantly associates with chronic rhinosinusitis disease severity. Allergy 2019;74:1569-72. [Crossref] [PubMed]
- Bellinghausen C, Rohde GGU, Savelkoul PHM, et al. Viral-bacterial interactions in the respiratory tract. J Gen Virol 2016;97:3089-102. [Crossref] [PubMed]
- Edouard S, Million M, Bachar D, et al. The nasopharyngeal microbiota in patients with viral respiratory tract infections is enriched in bacterial pathogens. Eur J Clin Microbiol Infect Dis 2018;37:1725-33. [Crossref] [PubMed]
- de Steenhuijsen Piters WA, Heinonen S, Hasrat R, et al. Nasopharyngeal Microbiota, Host Transcriptome, and Disease Severity in Children with Respiratory Syncytial Virus Infection. Am J Respir Crit Care Med 2016;194:1104-15. [Crossref] [PubMed]
- Rosenfeld RM, Piccirillo JF, Chandrasekhar SS, et al. Clinical practice guideline (update): Adult Sinusitis Executive Summary. Otolaryngol Head Neck Surg 2015;152:598-609. [Crossref] [PubMed]
- Goggin RK, Bennett CA, Bassiouni A, et al. Comparative Viral Sampling in the Sinonasal Passages; Different Viruses at Different Sites. Front Cell Infect Microbiol 2018;8:334. [Crossref] [PubMed]
- Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019;37:852-7. Erratum in: Nat Biotechnol 2019;37:1091. [Crossref] [PubMed]
- Zhang J, Kobert K, Flouri T, et al. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014;30:614-20. [Crossref] [PubMed]
- Bokulich NA, Subramanian S, Faith JJ, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 2013;10:57-9. [Crossref] [PubMed]
- Amir A, McDonald D, Navas-Molina JA, et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2017;2:ee00191-16. [Crossref] [PubMed]
- DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006;72:5069-72. [Crossref] [PubMed]
- Bokulich NA, Kaehler BD, Rideout JR, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome 2018;6:90. [Crossref] [PubMed]
- Faith DP. Conservation evaluation and phylogenetic diversity. Biological Conservation 1992;61:1-10. [Crossref]
- Martí JM, Martínez-Martínez D, Rubio T, et al. Health and Disease Imprinted in the Time Variability of the Human Microbiome. mSystems 2017;2:ee00144-16. [Crossref] [PubMed]
- Rosas-Salazar C, Shilts MH, Tovchigrechko A, et al. Differences in the Nasopharyngeal Microbiome During Acute Respiratory Tract Infection With Human Rhinovirus and Respiratory Syncytial Virus in Infancy. J Infect Dis 2016;214:1924-8. [Crossref] [PubMed]
- Borges LGDA, Giongo A, Pereira LM, et al. Comparison of the nasopharynx microbiome between influenza and non-influenza cases of severe acute respiratory infections: A pilot study. Health Sci Rep 2018;1:e47. [Crossref] [PubMed]
- Ding T, Song T, Zhou B, et al. Microbial Composition of the Human Nasopharynx Varies According to Influenza Virus Type and Vaccination Status. mBio 2019;10:01296-19. [Crossref] [PubMed]
- Berger A. Th1 and Th2 responses: what are they? BMJ 2000;321:424. [Crossref] [PubMed]
- König K, Klemens C, Haack M, et al. Cytokine patterns in nasal secretion of non-atopic patients distinguish between chronic rhinosinusitis with or without nasal polys. Allergy Asthma Clin Immunol 2016;12:19. [Crossref] [PubMed]
- Rosenberg HF, Dyer KD, Domachowske JB. Respiratory viruses and eosinophils: exploring the connections. Antiviral Res 2009;83:1-9. [Crossref] [PubMed]
- Rosenberg HF, Masterson JC, Furuta GT. Eosinophils, probiotics, and the microbiome. J Leukoc Biol 2016;100:881-8. [Crossref] [PubMed]
Cite this article as: Goggin RK, Bennett CA, Cooksley CM, Bassiouni A, Bialasiewicz S, Wormald PJ, Vreugde S, Psaltis AJ. Viral presence and the bacterial microbiome in chronic rhinosinusitis. Aust J Otolaryngol 2022;5:11.


